Use of Machine Learning Techniques for Case-Detection of Varicella Zoster Using Routinely Collected Textual Ambulatory Records: Pilot Observational Study
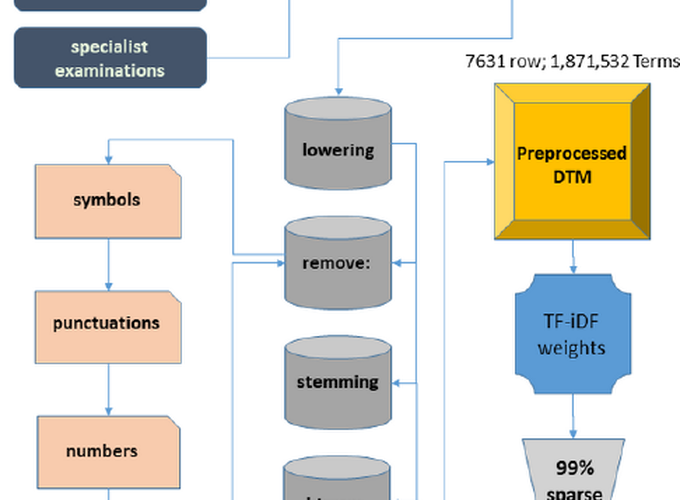 Flowchart from the acquisition, preprocessing, dimensionality reduction, and creation of the final DTM used (green).
Flowchart from the acquisition, preprocessing, dimensionality reduction, and creation of the final DTM used (green).
Use of Machine Learning Techniques for Case-Detection of Varicella Zoster Using Routinely Collected Textual Ambulatory Records: Pilot Observational Study
Abstract
Background: The detection of infectious diseases through the analysis of free text on electronic health reports (EHRs) can provide prompt and accurate background information for the implementation of preventative measures, such as advertising and monitoring the effectiveness of vaccination campaigns. Objective: The purpose of this paper is to compare machine learning techniques in their application to EHR analysis for disease detection. Methods: The Pedianet database was used as a data source for a real-world scenario on the identification of cases of varicella. The models’ training and test sets were based on two different Italian regions’ (Veneto and Sicilia) data sets of 7631 patients and 1,230,355 records, and 2347 patients and 569,926 records, respectively, for whom a gold standard of varicella diagnosis was available. Elastic-net regularized generalized linear model (GLMNet), maximum entropy (MAXENT), and LogitBoost (boosting) algorithms were implemented in a supervised environment and 5-fold cross-validated. The document-term matrix generated by the training set involves a dictionary of 1,871,532 tokens. The analysis was conducted on a subset of 29,096 tokens, corresponding to a matrix with no more than a 99% sparsity ratio. Results: The highest predictive values were achieved through boosting (positive predicative value [PPV] 63.1, 95% CI 42.7-83.5 and negative predicative value [NPV] 98.8, 95% CI 98.3-99.3). GLMNet delivered superior predictive capability compared to MAXENT (PPV 24.5% and NPV 98.3% vs PPV 11.0% and NPV 98.0%). MAXENT and GLMNet predictions weakly agree with each other (agreement coefficient 1 [AC1]=0.60, 95% CI 0.58-0.62), as well as with LogitBoost (MAXENT: AC1=0.64, 95% CI 0.63-0.66 and GLMNet: AC1=0.53, 95% CI 0.51-0.55). Conclusions: Boosting has demonstrated promising performance in large-scale EHR-based infectious disease identification.